Według Międzynarodowej Federacji Diabetologicznej (IDF) na cukrzycę choruje ponad 400 milionów ludzi na całym świecie, a w samej Polsce obserwuje się tendencję wzrostową z 2,8 milionami pacjentów. Cukrzyca charakteryzuje się nieprawidłowym obrotem glukozy w organizmie, a jej poziom w surowicy krwi jest kontrolowany przez skoordynowane działanie hormonów trzustkowych. Jeden z takich hormonów, zwany insuliną, wytwarzany przez komórki β wysp trzustki, jest odpowiedzialny za wychwyt glukozy i przekaz sygnału do innych komórek organizmu. Zatem niewystarczająca liczba komórek β w organizmie lub ich nieprawidłowe działanie prowadzi do stanu z podwyższonym poziomem glukozy, zwanego hiperglikemią, co z kolei prowadzi do cukrzycy. Częstość występowania cukrzycy w świecie zachodnim wzrasta z powodu diety i stylu życia, a wiek nowo zdiagnozowanych pacjentów systematycznie się obniża. Pomimo tego, że cukrzyca jest uważana za chorobę metaboliczną, większość dotychczasowych badań dotyczyła globalnych zmian w genomie (na poziomie DNA), transkryptach (na poziomie RNA), kilka badań odnosiło się do roli białek, a tylko nieliczne dotyczyły zmian w metabolitach (np. lipidach, cukrach czy aminokwasach). Jak dotąd nie przeprowadzono jeszcze kompleksowej analizy ilościowej na różnych poziomach organizacji komórki. Aby wypełnić te luki, zaproponowaliśmy kompleksowe badania skupiające się na zmianach na poziomie RNA (globalna analiza transkryptomu), proteomie (analizy globalne i mitochondriów) i metabolomie (analizy globalne i specyficzne badania metabolitów), w celu wskazania różnic w zmianach związanych z rozwojem ludzkich komórek β i cukrzycą. Aby osiągnąć ten cel, zastosujemy model różnicowania się komórek trzustki z wykorzystaniem ludzkich indukowanych pluripotencjalnych komórek macierzystych, które są zdolne do powielenia rozwoju komórek β trzustki in vitro. Do badań wybraliśmy również specyficzną formę cukrzycy, ze skłonnością do ketozy (forma KPD), i będziemy wykorzystywać indukowane pluripotencjalne komórki macierzyste pochodzące od pacjenta z tą postacią choroby. Realizując cele projektu spodziewamy się, że wykonując kompleksowe analizy multiomiczne na różnych poziomach organizacji komórkowej, odkryjemy specyficzne scieżki przekaźnictwa sygnału odpowiedzialne za rozwoj komórek β trzustki. Planujemy wykorzystanie tego samego repertuaru technik w połączeniu z różnymi analizami czynnościowymi komórek β w celu zdefiniowania patogenezy molekularnej cukrzycy i wskazania nowych potencjalnych celów terapeutycznych dla przyszłych procedur klinicznych i wdrażania leków. Projekt ten będzie realizowany w instytucji goszczącej (Uniwersytet im. Adama Mickiewicza w Poznaniu), w laboratorium kierowanym przez prof. Małgorzatę Borowiak, która posiada rozległą wiedzę specjalistyczną w zakresie badań nad cukrzycą, wykorzystującą innowacyjne metody różnicowania ludzkich pluripotencjalnych komórek macierzystych w kierunku trzustki. Kierownik wniosku, docent dr hab. Maciej M. Łałowski specjalizuje się w technikach spektrometrii mas i jest ekspertem w zakresie wieloetapowej multiomicznej integracji danych. Łącząc wiedzę ekspercką zarówno kierownika projektu, jak i laboratorium przyjmującego, spodziewamy się szczegółowo opisać mechanizmy komórkowe leżące u podstaw cukrzycy.
About project
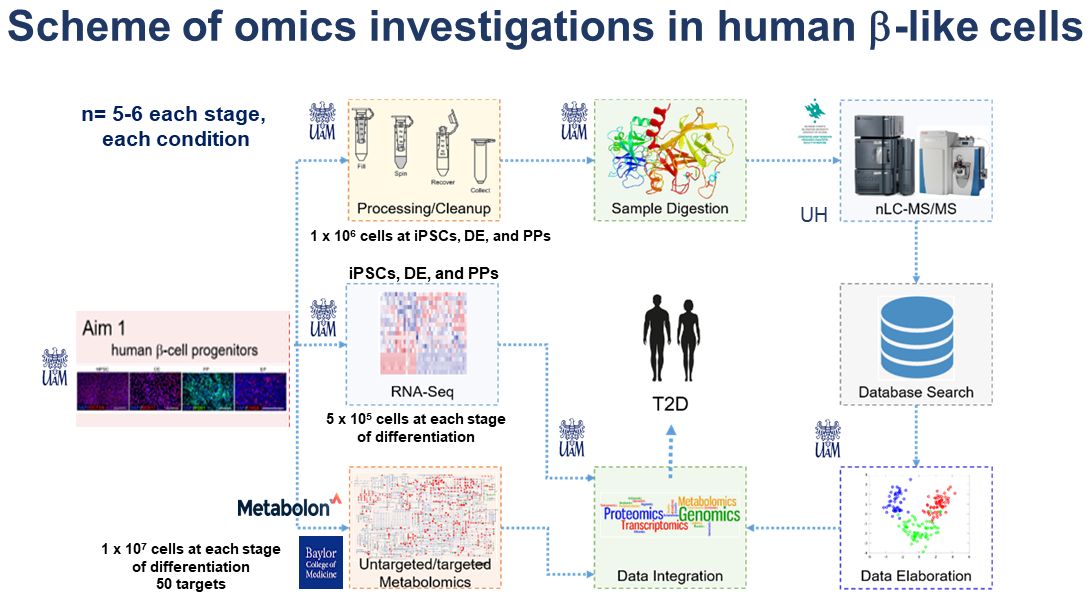
Diabetes is a metabolic disease characterized by abnormal glucose homeostasis. The glucose levels in blood serum are regulated by coordinated action of pancreatic hormones, with insulin being responsible for glucose uptake by cells in the periphery. The sole source of insulin in humans are pancreatic islet β-cells. Therefore, the insufficient β-cell number or function leads to hyperglycemia, which in turn might develop into diabetes. Diabetes is worldwide burden, affecting more than 400 million of people according to the International Diabetes Federation, and in Poland alone displays an upward trend with 2.8 million (2018). The insulin resistance in the periphery is an additional factor contributing to diabetes severeness, however, more evidence points to β-cells being the key factor in the disease. Multiple factors might affect β-cell number and health as highlighted by various studies over the years. The comprehensive quantitative multiomic analysis at various levels of organization (i.e., global transcriptome, proteome, and metabolome) has not yet been performed. In this proposal, we postulate a multiomic study of transcriptome, proteome, and metabolome to pinpoint the differential changes associated with human β-cell development (Aim 1), and disease (Aim 2). To achieve it, we propose to employ the human induced pluripotent stem cell (hiPSC) pancreatic differentiation model, enabling the recapitulation of the human β-cell development in vitro in stepwise manner, reflecting the key developmental events. Based on previous literature we hypothesize that various stages of development of human β-cells are associated with significant changes at different levels of organization of the cellular “-ome”. By performing the comprehensive multi-level omics investigations, we are expecting to reveal the differentially expressed genes (by RNA-seq), proteins with differential abundance (label-free quantitative proteomics) and differentially regulated metabolites (untargeted metabolomics) and finally the signaling cues behind the development of β-cells. Furthermore, to model human diabetes in vitro, we chose the ketosis prone (KPD) type of diabetes, and we will utilize the hiPSCs derived from a patient with KPD. We have previously performed genetic analysis coupled with preliminary multiomic investigations in β-cells derived from a KPD patient with an unusual clinical course and a potentially functional variant in the PDX1 gene and controls. During differentiation of this patient’s iPSCs through defined cellular stages of the pancreatic lineage, we were able to recapitulate the Leucine hypercatabolism common in KPD patients in pancreatic cells in vitro. In the second aim of the project, we propose to extend these findings, by combining the power of multi-level omics approach (on whole cells and isolated mitochondria), and following the treatment with a common diabetic drug, metformin with rigorous functional assessment in KPD patients’ cells. For comprehensive multiomics integration, in addition to selected bioinformatic platforms we will utilize fuzzy c‐means clustering, in which transcripts, proteins, and metabolites will be assigned clusters based on their abundance patterns and enriched gene ontology and KEGG pathways terms, to pinpoint differentially altered pathways and functional network modules suitable for subsequent functional validations.
With new lines of personalized medicine emerging, there is a growing demand to understand the specific disease mechanisms as well as to identify the prospective biomarkers to stratify patients and to monitor their treatment. The use of multiple omics technologies towards various forms of diabetes carries the potential to address such needs. This project will be performed in close collaboration with the host laboratory of Prof. Malgorzata Borowiak and her team at the Adam Mickiewicz University- Poznan, Poland. She is a world-leading expert in hiPSCs differentiation towards pancreatic β-cells as well as the β-cell biology and function, including diabetes molecular modeling. By combing the expertise of the host institution with the multiomics data integration expertise brought by the PI we propose to extend the available preliminary data and analyze the global changes at RNA, protein and metabolite level in control and patient developing pancreatic cells to build a dynamic map of changes in type 2 diabetes (T2D).

Research group information

With new lines of personalized medicine emerging, there is a growing demand to understand the specific disease mechanisms as well as to identify the prospective biomarkers to stratify patients and to monitor their treatment. The use of multiple omics technologies towards various forms of diabetes carries the potential to address such needs. This Polonez Bis project is realized in close collaboration with the host laboratory of Prof. Malgorzata Borowiak and her team at the AMU. By combing the expertise of the host institution with multiomics data integration expertise brought by the PI we propose to analyse the global changes at RNA, protein and metabolite level in control and patient developing pancreatic cells to build a dynamic map of changes in type 2 diabetes (T2D). Our main approaches are based on NGS (RNAseq), combined with mass spectrometry. We use both experimental and computational approaches.
Group members information

dr hab. Maciej Maurycy Łałowski
PI / visiting professor
maciej.lalowski@amu.edu.pl
+48 61 829 5855
orcid.org
Host, prof. Małgorzata Borowiak
AMU professor
malgorzata.borowiak@amu.edu.pl
+48 61 829 5965
orcid.org
Maciej Lalowski is an experimental and computational biologist. He obtained his PhD at the Polish Academy Sciences in Warsaw/NYU Medical Center, NYC, USA. Maciej did hist postdoctoral training at the University of Helsinki, Max Planck Institute for Molecular Genetics and Max Delbrűck Center for Molecular Medicine in Berlin Germany. He has been awarded docentship at the University of Helsinki (in 2011), and specializes in quantitative mass spectrometry and systems biology. He is the current president of the Finnish Proteomics Society. Maciej has effectively applied omics methodologies to study various aspects of neurodegeneration, muscle aging, heart transplantation and development, cancer cachexia among others, to reveal affected pathways and network modules, suitable for future therapeutic interventions.
Maciej has been awarded a Polonez Bis 1 grant to set up his own research group in November 2022, hosted by Prof. Borowiak at AMU. In his spare time Maciej loves biking and listening to good music.

dr Artur Jankowski
Lab manager / research assistant
artur.jankowski@amu.edu.pl
+48 618 29 5961
orcid.org
Artur is an experimental biologist and a graduate of the University of Adam Mickiewicz in Poznań, where he obtained a master’s degree in experimental biology. Then he continued his doctoral thesis at the University in cooperation with the Institute of Dendorology of the Polish Academy of Sciences in Kórnik. He received his PhD degree in 2021. Artur has been working as a laboratory manager in the Stem Cells Laboratory (prof. Borowiak’s group) for four years and he makes sure that the functioning of the laboratory runs smoothly.

MSc Edyta Urbaniak
PhD student
edyta.urbaniak@amu.edu.pl
+48 618 29 5854
Edyta is a biotechnologist, a graduate of the University of Adam Mickiewicz in Poznań. She obtained her master’s degree in biotechnology in UAM in the stem cell laboratory, where she then worked as a contractor on a project related to the maturation of the pancreas, currently she is a PhD student working on a project related to the maturation of human insulin. During these years, she gained experience in working with stem cell culture and various techniques of molecular biology. Within the context of Polonez Bis project, Edyta will perform differentiation of iPSCs and pancreatic cells obtained from patients with atypical diabetes using advanced microscopic imaging and focused functional analyses.

lic Maja Bagińska
MSc student
majbag@st.amu.edu.pl
+48 618 29 5961
Maja is pursuing her Master’s degree in Biotechnology at the Adam Mickiewicz University. During her studies, she was involved in researching the mechanisms of insulin folding in the pathogenesis of diabetes. The studies were conducted in the Stem Cell Laboratory, AMU under the supervision of Prof. Małgorzata Borowiak. As a part of a grant project, ID-UB 118 Study@research (Inicjatywa Doskonałości – Uczelnia Badawcza, AMU) Maja is also involved in characterization of the early stages of differentiation process of pancreatic cells with the OVOL2 gene knockout. Within the context of Polonez Bis project, Maja will perform differentiation of iPSCs and pancreatic cells obtained from patients with atypical diabetes using microscopic imaging and preliminary functional analyses.
Alumni

MSc Maroua Rami
Within the context of Polonez Bis project, Maroua has performed the initial multi-omics characterization of differentiating healthy pancreatic β-cells
Events
Seminar: Use of multi-omics in disease studies, 19.11.2024
Maciej Łałowski, Adam Mickiewicz University in Poznań
Maria Skłodowska-Curie National Center of Oncology, branch in Gliwice, Poland
Host: Prof. Monika Pietrowska
Invited lecture: Multi-omics in development and disease – heart and rare diabetes, 22.11.2024
Maciej Łałowski, Adam Mickiewicz University in Poznań
XXVIII Gliwice Scientific Meetings, 2024, https://gsn.io.gliwice.pl/index.php/programme
Public audience lectures
Lecture: Zastosowanie technik omicznych do badania zdrowych i chorych komórek beta trzustki i wydzielania insuliny
- Zespół Szkół Ogólnokształcących w Krzyżu Wielkopolskim, ul. Sienkiewicza 1 on 29.10.2024
- II Liceum Ogólnokształcące im. Generałowej Zamoyskiej i Heleny Modrzejewskiej, 1349 IB school, ul. Matejki 8/10 on 31.10.2024
DEM-Aging: News for the two faces of dementia, 13th June 2024
IRCCS Fondazione Stella Maris, Calambrone (Pisa), Italy
Dr. Maciej Lalowski
Multi-omics towards complex diseases: lessons to be learned
Adam Mickiewicz University, Poznań and University of Helsinki, Finland
Seminar „Utilization of multiomics approach in the studies of human diseases” given by dr hab. Maciej Łałowski – dr hab. Maciej Łałowski from Department of Gene Expression, IBMiB, Adam Mickiewicz University.
The lecture is organized as part of the KNOW RNA Research Centre in Poznań and RNA Society – RNA Salon Poznań and will be held in person on the 13 th of January at 9:15, in the Aula Paczoskiego, Faculty of Biology AMU.
GOSH / GGM Metabolic Meeting (UCL-London, UK):
Wednesday 29 March 2023 at 12 noon as a Zoom meeting.
Prof Maciej M Lalowski
Maciej Maurycy Lalowski works at the Department of Gene Expression, IMBiB, Adam Mickiewicz University, Poznan/Poland and is affiliated to Medicum, Department of Biochemistry and Developmental Biology, Meilahti Clinical Proteomics Core Facility, HiLIFE, University of Helsinki-Finland. Maciej does research in Proteomics, Neurodegeneration and Mass Spectrometry Imaging. The current research projects relate to Diabetes, Alzheimer’s Disease, Mood Disorders, Neuronal Ceroid Lipofuscinosis and Schizophrenia.
Compartmental proteomics reveals mechanisms behind the pathogenesis of CLN5 disease
Polish Proteomics Society
Scientific Meeting 8th December 2023
Institute of Bioorganic Chemistry PAS, Poznań
Dr. Maciej Lalowski
Multi-omics towards disease: case of neonatal heart and atypical diabetes
Adam Mickiewicz University, Poznań and University of Helsinki, Finland
Selected publications
- Laakkonen E, Soliymani R, Rintala P, Sipilä S, Kaprio J, Kujala U, Baumann M, Kovanen V, Lalowski M. Proteomic analysis of skeletal muscle reveals estrogen as a key regulator of muscle signaling in women. Aging Cell. 2017 Dec;16(6):1276-1287. doi: 10.1111/acel.12661
- Hulmi JJ, Penna F, Pöllänen N, Nissinen TA, Hentilä J, Lautaoja JH, Ballarò R, Soliymani R, Baumann M, Ritvos O, Pirinen E, Lalowski M. Increased muscle Serpina3n and NAD+depletion are molecular determinants of cancer cachexia – the effects of blocking myostatin/activins. 2020, Mol Metabolism, 2020 Jun 26;41:101046. doi: 10.1016/j.molmet.2020.101046
- Kankuri E, Finckenberg, Leinonen J, Tarkia M, Björk S, Purhonen J, Kallijärvi J, Kankainen M, Soliymani R, Lalowski M, Mervaala E. Altered acylcarnitine metabolism and inflexible mitochondrial fuel utilization characterize the loss of neonatal myocardial regeneration capacity. Exp Mol Medicine, 2023, . doi: 10.1038/s12276-023-00967-5
- Scavuzzo MA, Borowiak M. Two drugs converged in a pancreatic β cell. 2020, Science Translational Medicine 12;530. doi: 10.1126/scitranslmed.aba7359
- Yang D, Patel S, Szlachcic WJ, Chmielowiec J, Scaduto D, Putluri N, Sreekumar A, Suliburk J, Metzker M, Balasubramanyam A, Borowiak M. Pancreatic Differentiation of Stem Cells Reveals Pathogenesis of a Syndrome of Ketosis-Prone, 2021, Diabetes 70;10;2419-2429. doi: 10.2337/DB20-1293
- Chmielowiec J, Szlachcic WJ, Yang D, Scavuzzo MA, Wamble K, Sarrion-Perdigones A, Sabek OM, Venken K.JT, Borowiak M. Human pancreatic microenvironment promotes β-cell differentiation via non-canonical WNT5A/JNK and BMP signaling, 2022, Nature Communications, 13;1;1952. doi: 10.1038/s41467-022-29646-1
- Sikorski V, Selberg S, Lalowski M, Karelson M, Kankuri E.The structure and function of YTHDF epitranscriptomic m6A readers. Trends Pharmacol Sci. 2023 Jun;44(6):335-353. doi: 10.1016/j.tips.2023.03.004
- Urbaniak E, Henry S, Lalowski M and Borowiak M (2025) Molecular puzzle of insulin: structural assembly pathways and their role in diabetes. Front. Cell Dev. Biol. 13:1502469. doi: 10.3389/fcell.2025.1502469 (supported by Polonez Bis grant).